Chapter 9 Spatial Estimation
Written by Sushil Nepal and Paul Pickell
Geostatistics uses the metrics based on statistical tools to characterize the distribution of a random variable across a geographical region of interest (Getis et al. 2004). Like any other data, spatial data may be incomplete, which can limit our analytical capacity in the incomplete regions. In context of spatial data, we wish to understand spatial phenomena like events or processes that occur over large areas, but usually we are limited in our capacity to sample everywhere. Most of the time, only discrete and unrepresentative information on the spatial phenomenon is collected, which does not allow us to create a precise and continuous map of the phenomenon over entire geographic area of interest. How can we best utilize the available samples to represent the phenomenon across the entire geographic area? This chapter introduces some basic ideas on different types of sampling strategies in spatial context. In addition, this chapter also introduces various type of spatial statistics that are available to predict the occurrence of spatial phenomena in unsampled locations.
- Differentiate the advantages of spatial sampling strategies
- Describe the relationship between observations at different spatial scales using autocorrelation and semivariogram
- Apply methods of spatial interpolation to predict observations at unknown locations
- Apply methods of spatial prediction using regression models
9.1 Spatial Autocorrelation
A key characteristic that distinguishes spatial data from other types of data is the fact that spatial phenomena are frequently spatially autocorrelated. Spatial autocorrelation is the relationship between a variable of interest with itself when measured at different locations (Cliff 1973). Any two samples are more likely to be correlated or similar to each other when the samples are closer in space compared with two samples that are taken at farther distances. For example, if we measured air temperature at our current location and then walked 1 m away and measured it again, then walked 10 km and measured it again, we would expect that the temperatures sampled 1 m apart would be more similar than the temperatures measured 10 km apart. The precise degree and sign (positive or negative) of spatial autocorrelation will vary from phenomenon to phenomenon because some phenomena change more quickly over space than others. As far as we can tell, all spatial phenomenon exhibit spatial autocorrelation at some, but not all distances. In other words, we have never observed a natural spatial phenomenon that is truly random. Spatial autocorrelation provides a tremendous advantage for estimating statistics and characteristics of populations in space, but also introduces several important caveats.
Consider the following example; Figure 9.14 shows the clustering pattern in the given square boxes (can be variable of interest) representing the positive spatial autocorrelation (left) and a complete checkerboard (right) distribution of square boxes (variable of interest) indicating a negative spatial autocorrelation.

Figure 9.1: Example of a positive (left) and negative spatial autocorrelation (right) for a give domain. Nepal, CC-BY-SA-4.0.
Example of a positive (left) and negative spatial autocorrelation (right) for a give domain.
9.4 Moran’s I
Moran’s I (Moran 1950), is a correlation coefficient that measures the degree of spatial autocorrelation in certain attributes of the data. It is based on the spatial covariance standardized by the variance of the data (Moran 1950). It works based on the neighborhood list created based on spatial weight matrix (Suryowati, Bekti, and Faradila 2018). The value of Moran’s I ranges between -1 to 1, where 1 indicates the perfect positive spatial autocorrelation, 0 indicates the random pattern, and -1 indicates the perfect negative autocorrelation (Moran 1950). Moran’s I is calculated using the following formula (Moran 1950):
\[ I= \frac{1}{s^2} \frac{\sum_{i}\sum_{j}({y_i-\bar{y})({y_j-\bar{y}})}} {\sum_{i}\sum_{j}w_{ij}} \] Where, \[I=\text{the Moran I statistics}\], \[y_i=\text{variable measure at location i}\] \[y_j=\text{variable measure at location j}\]
\[S^2=\text{the variance}\] \[w_{ij}=\text{the spatial weight matrix}\]
9.5 Case Study 1
For this case study, we will use ground plot data from Change Monitoring Inventory (CMI) program (for details: Province of BC 2018) for Williams Lake and 100-miles House timber supply area (TSA) in the province of British Columbia, Canada. William Lake TSA and 100-miles House TSA are divided into 18 and 8 blocks respectively Figure 9.1. There is a total of 456 CMI plots used in this study Figure 9.1. The total basal area (m2/ha) is our variable of interest in this study. For each of the polygon in Williams lake and 100-miles house TSA, total basal area was calculated by taking the sum of the basal area of each CMI plots in each polygon. For this part of exercise, we want to understand if there is any spatial relationship (autocorrelation) between the total basal area measured in each polygon of TSA. We will quantitatively measure the presence or absence of spatial autocorrelation using Moran’s I and Geary’s C.
## OGR data source with driver: ESRI Shapefile
## Source: "C:\Users\paul\Documents\geomatics-textbook\geomatics-textbook\data\10", layer: "Block_basa_area"
## with 26 features
## It has 21 fields
## Integer64 fields read as strings: Field1## OGR data source with driver: ESRI Shapefile
## Source: "C:\Users\paul\Documents\geomatics-textbook\geomatics-textbook\data\10", layer: "100_and_will"
## with 2 features
## It has 16 fields
Figure 9.2: CMI plots location for williams lake TSA and 100 miles house TSA. Basal area has been calculated using the sum of the basal area for each plot (dot) over the given polygon. Nepal, CC-BY-SA-4.0.
CMI plots location for williams lake TSA and 100 miles house TSA. Basal area has been calculated using the sum of the basal area for each plot (dot) over the given polygon. :::
Calculating Moran’s I
We will calculate the Moran’s I for the basal area variable pertaining to the polygon.
Using Contiguity
Define Neighborhood
The Moran’s I statistic is the correlation coefficient for the relationship between a variable and its surrounding values. But before we go about computing this correlation, we need to come up with a way to define a neighborhood. There are two ways to define neighborhood namely; contiguity for spatial polygon data and distance-based approach for the spatial point data and polygon data both. For polygon data, contiguity based neighborhood selection can be adopted using two widely used method, respectively known as Rook’s case or Queen’s case Figure 9.2.

Figure 9.3: Rook’s (left) and Queen’s (right) case for searching the neighborhood (grey unit) for the darker unit in the center. Nepal, CC-BY-SA-4.0.
Rook’s (left) and Queen’s (right) case for searching the neighborhood (grey unit) for the darker unit in the center.
Step 1: Build Neighborhood
Since we are working with the polygons, we will use the queen’s contiguity to build the neighborhood list using polyn2b function in spdep package in R and plot the the linkage**
####################### Plot the data #######################
### Convert the spatial data into data frame ##########
filename_df <- tidy(filename)
# make sure the shapefile attribute table has an id column
filename$id <- rownames(filename@data)
# join the attribute table from the spatial object to the new data frame
filename_df <- left_join(filename_df,
filename@data,
by = "id")
#Searching neighborhood
w1 <- poly2nb(filename,row.names=filename$id,queen=T) ###### queens case
coords <- coordinates(filename)
plot(w1, coords, col="grey")
Step 2: Getting a Spatial Weight Matrix for Neighborhood List
Step 3: Calculate Moran’s Correlation Coefficient Using the Spatial Weight Matrix for Neighbors
## $I
## [1] -0.1112932
##
## $K
## [1] 4.854083Step 4: Conduct the Significance Test for the Calculated Moran’s I Value
##
## Moran I test under randomisation
##
## data: filename$Basal
## weights: ww
##
## Moran I statistic standard deviate = -0.55645, p-value = 0.711
## alternative hypothesis: greater
## sample estimates:
## Moran I statistic Expectation Variance
## -0.11129321 -0.04000000 0.01641516Using the K-Nearest Neighborhood Imputation
Step 1: We will select 3 Nearest Neighbor Using Distance-based Approach
The function knearneigh in ’spdep” package in R and build a spatial weight
# Searching neighborhood
col.knn <- knearneigh(coords, k=3)
w<-knn2nb(col.knn,row.names = filename$id)
coords <- coordinates(filename)
plot(w, coords, col="grey") Step 2: Build a Spatial Weight Matrix for the Neighborhood List
Step 2: Build a Spatial Weight Matrix for the Neighborhood List
Step 3: Calculate the Moran’s I Coefficient
## $I
## [1] 0.01767042
##
## $K
## [1] 4.854083Step 4: Significance Test for Calculated Moran’s I
##
## Moran I test under randomisation
##
## data: filename$Basal
## weights: ww1
##
## Moran I statistic standard deviate = 0.43976, p-value = 0.3301
## alternative hypothesis: greater
## sample estimates:
## Moran I statistic Expectation Variance
## 0.01767042 -0.04000000 0.01719815Note that the value of Moran’s I changed based on how we calculated the neighborhood list using two different approach. The interpretation change here based on the way we created the neighborhood. With contiguity based neighbor, we found a negative value for I (-0.11), indicating a negative weak spatial autocorrelation. When we run the significance test we can see that the p-value < 0.05 indicating the autocorrealtion is not significant. While using nearest neighbor we found that I (0.017) indicated a weak positive spatial autocorrelation. One reason for the difference is k-nearest neighbor uses polygon within a greater distance and can include more polygons as compared to contiguous neighbor which uses either “queens” or “rooks” contiguity (Suryowati, Bekti, and Faradila 2018).
9.6 Geary’s C
Another more local measure of spatial autocorrelation unlike Moran’s I is Geary’s C (Geary 1954). While Moran’s I is calculated by standardizing the spatial autocovariance by the variance of the data. Geary’s c on the other had uses the sum of the squared differences between pairs of data values as it is a measure of covariance (Geary 1954). However, both statistics depends on the spatial nature of data and are based on neighborhood. Both of these statistics depend on a spatial structure specified by a spatial weights matrix. The value of Geary’s C ranges between 0 to some unknown positive value, where 0 indicates the spatial randomness, values less than 1 indicates the positive spatial autocorrelation, while value greater than 1 indicates negative spatial autocorrelation (Geary 1954). It is calculated using following formula: \[ C=\frac{(n-1) \sum_{i}^n\sum_{j}^nw_{ij}(y_i-y_j)^2}{2\sum_{i}^n\sum_{j}^nw_{ij}\sum_{i}(y_{i}-\bar{y})^2}\]
Step 1 and 2: We will calculate all the neighborhood list exactly how we did for Moran’s I and get our spatial weight matrix
Step 2: In our final step, we will use geary funtion from “spdep” package to calculate the value of Geary’s
For Queens Case
## Geary C
geary(filename$Basal, ww, n=length(ww$neighbours),n1=length(ww$neighbours)-1, S0=Szero(ww))## $C
## [1] 0.9944114
##
## $K
## [1] 4.854083##
## Geary C test under randomisation
##
## data: filename$Basal
## weights: ww
##
## Geary C statistic standard deviate = 0.02595, p-value = 0.4896
## alternative hypothesis: Expectation greater than statistic
## sample estimates:
## Geary C statistic Expectation Variance
## 0.99441140 1.00000000 0.04638142For Nearest Neighbour Method
## Geary C
geary(filename$Basal, ww1, n=length(ww1$neighbours),n1=length(ww1$neighbours)-1, S0=Szero(ww1))## $C
## [1] 0.9416159
##
## $K
## [1] 4.854083##
## Geary C test under randomisation
##
## data: filename$Basal
## weights: ww1
##
## Geary C statistic standard deviate = 0.3791, p-value = 0.3523
## alternative hypothesis: Expectation greater than statistic
## sample estimates:
## Geary C statistic Expectation Variance
## 0.94161588 1.00000000 0.02371762Note that the value of Geary’s C indicated a positive spatial autocorrelation using both queens case and k-nearest neighbor. However, the spatial autocorrelation was not significant as given by p-value < 0.05. Both Moran’s I and Geary’s C are in agreement in terms of results.
9.7 Populations, Samples and Statistics
Suppose you are interested in measuring the heights of trees in a forest. There are a lot of trees in this forest and you cannot measure all of them. So you cleverly decide to sample some locations and infer the mean tree height of the forest from this sample. In this example, the number of trees that you measured are the sample \(n\), the forest is the total population of trees \(N\), and the statistic that you are interested in estimating is the mean tree height of the forest:
\[ x̄ = \frac{1}{n} \sum_{i=1}^{n} x_i \] Lowercase \(x\) represents any sampled tree and \(x̄\) represents the mean of the sample, which is an estimate of the population mean. It is important to recognize that statistics of samples and populations are discussed and represented differently. The true mean tree height of the forest is denoted by the Greek lowercase mu \(μ\), and we can never know this value without measuring every tree. So instead, we infer the population statistic using the sampled data and hope that the sample statistic will be close to the true population statistic, but we must accept that they are rarely equivalent \(x̄≠μ\). The same equation above can be re-written for the population mean:
\[ μ = \frac{1}{N} \sum_{i=1}^{N} X_i \]
Notice here we are using \(μ\) to signify the population mean tree height, uppercase \(N\) signifies all the trees in the population, and uppercase \(X_i\) is one tree in the population. At this point, you might take a moment to appreciate that as our sample \(n\) approaches \(N\), our sample mean \(x̄\) should also approach the population mean \(μ\). In other words, the more trees we sample, the more likely we are going to have an accurate estimate of \(μ\). It might surprise you that this conclusion is likely true for large samples \(n\), but not small samples \(n\). Why? Because any randomly-selected tree from our forest \(n=1\) could be much taller or shorter than the mean just due to random chance alone. In fact, if our forest has two distinct cohorts of trees, one very young emerging below the canopy and one established in the overstory (i.e., bi-modal distribution), then the population mean might never be approximated by any particular tree. This is problematic for us, because we want to accurately estimate the population statistic whilst exhausting as few of our resources as possible to do so (i.e., time, people, equipment, budget).
When we made our measures of tree heights, it would be reasonable for us to expect that the tree heights that we sampled are going to be different (i.e., most trees will not have the same height). The magnitude of those differences in the sample are captured by the variance, which is a measure of the dispersion of our tree heights \(x\) relative to the sample mean \(x̄\):
\[ s^2 = \frac{1}{n} \sum_{i=1}^{n} (x_i-x̄)^2 \] We square the differences so that the variance is always positively signed and consequently variance always has squared units, hence \(s^2\). If we take the square root of the variance, we have the standard deviation:
\[ s = \sqrt{s^2} \] The population standard deviation is represented by Greek lowercase sigma \(σ\) and population variance is \(σ^2\).
9.8 Significance Testing
The significance level of a test statistic from the sample is compared with the critical value of the test statistic obtained by repeatedly reordering the data with random proportions (Jacquez 1999). Interpretation of this significance level in classical statistics is done using a p-value which, if less than or equal to a defined threshold, also know as alpha α (usually 5% or 0.05), then the null hypothesis of “no difference from random” is rejected (Jacquez 1999). In other words, when the p-value is less than or equal to 0.05, we conclude that the statistic we observe for the sample is unlikely to have occurred due to random chance alone. This interpretation is important because recall that if our sample is small, there is a random chance that any statistic we calculate from it could be due to that particular sample rather than represent a true characteristic of the larger population. For example, imagine the case where we randomly sample some trees, but by chance we happen to only sample young trees and so our mean tree height from the sample is not representative of the population. The p-value represents the probability that our statistic from the sample is observed due to random chance, so we want this value to be as low as possible.
While \(α=0.05\) is widely used, there are cases where we may set a higher standard. For example, where the risk of incorrectly rejecting the null hypothesis, known as a “false positive” or Type I error, could have significant consequences on human health or safety. Consider the case where a new drug is being tested for efficacy and the p-value in the clinical trial is 0.05, so the null hypothesis that placebo is better than the drug is rejected. But there is a 5% probability that there is no difference between the new drug and a placebo. The implication is that 5% of the human population who take the new drug will not experience any benefits. If the population eligible for this drug is large, this can translate to many people being impacted by this decision. You can see how this can be problematic because there might be alternative therapies or drugs that are more effective than this new drug that the population are not utilizing. A similar case might exist where we believe that the null hypothesis is true, but it is in fact false, known as a “false negative” or Type II error. In these cases, we want to be sure that our result is not spurious, so the p-value standard may be raised to 0.01 or even 0.001, so that only one person in a hundred or one person in a thousand will experience no difference from the placebo.
9.9 Classical Statistics vs. Geostatistics
In classical statistics, the variance is assumed to be totally random when samples are drawn from a defined population (Jacquez 1999) and are assumed to come from one distribution (Steel and Torrie 1980). Inferences about the population can be made by comparing a statistic calculated for the sample to the distribution of that statistic under the null hypothesis for the assumed distribution (Jacquez 1999).
In geostatistics, the variance of a spatial phenomenon is assumed to be partly random and each point in the field represents a sample from some distribution (Jacquez 1999). However, the distribution at any one point may differ completely from all other points in its shape, mean, and variance. The distribution of differences in sample values separated by a specified distance is assumed to be the same over the entire field. Sample values that exhibit spatial autocorrelation due to their proximity to each other will have relatively small random variance of the distribution of differences. If the sample values do not exhibit spatial autocorrelation, then the variance is larger. The semivariance or half of the variance is used to measure the similarity between points at a given distance apart. The distance at which we compare the semivariance of sample values is known as the lag distance. If we graph semivariance and lag distance, we have created a semivariogram. Since we are usually dealing with samples and not the full population, we will almost never know the exact semi-variogram, instead we must rely on fitting models to estimate this relationship.
Thus, geostatistics is distinguished from classical statistics by the fact that we need to estimate the semivariogram function and then incorporate the semivariogram function to estimate the values at unsampled locations using various predictive spatial methods.
9.10 Semivariogram Modeling
The semivariogram is a basic geostatistical tool for measuring spatial autocorrelation of a variable measured at different spatial location. A semivariogram is a measure of variance of a variable between two locations separated by certain lag distance (Isaaks and Srivastava 1989). For example, we can measure how a variable \(y\) changes in value between sites \(i\) and \(j\) by calculating the variance between the sites \(y_i-y_j\). If the surface represented by the two points is continuous and the lag is a small distance, we would expect the variance to be small (Isaaks and Srivastava 1989). With increasing lag distance, we would expect the variance to increase. Let us translate this intuitive statement into a equation known as the empirical variogram:
\[\gamma{(h)}=\frac{1}{2N}\sum_{i,j=1}^{N(h)}{({y_i}-y_j)}^2\]
Where, \[\gamma{h}=\text{semivariance at a spatial lag h}\] \[ i=\text{measure spatial coordinate (latitude/UTM easting)}\] \[ j=\text{measure spatial coordinate (longitude/UTM northing)}\] \[y_{i}=\text{measured value of variable of interest at the spatial location i} \] \[y_{j}=\text{measured value of variable of interest at the spatial location j} \] \[N=\text{number of sampled differences or lag} \]
Like the familiar variance, it is a sum of squared differences divided by the number \(N\) of sampled differences. Unlike simple variance about a mean that we discussed earlier, the semivariogram measures difference between two sample values. The semi in semivariogram comes from the fact that the variance is divided in half.
A semivariogram is a graph that consists of semivariance on the y-axis and a lag distance on the x-axis Figure 9.4. There are some important features of a semivariogram that can be used to interpret the nature and structure of spatial autocorrelation.

Figure 9.4: An example semivariogram with all the components using the ‘Fulmar’data from ’gstat’ package in R. Nepal, CC-BY-SA-4.0.
Range \(a\): The distance at which a variogram model first flattens out. This is the distance up to which the variable is considered to be spatially autocorrelated (Figure 9.4). For variogram models that are bounded and have sills, the range is generally accepted to be 95% of the sill.
Nugget \(c_0\): Nugget refers to unaccounted autocorrelation due to a smaller lag distance than sampling distance or due to errors and imprecision arising from sampling (Figure 9.4). The nugget is also where the variance function \(\gamma{(h)}\) intercepts the y-axis. Without the nugget, we would expect all variogram models to evaluate to zero variance at zero lag \(\gamma{(0)}=0\), and conceptually this makes sense because how can you have variance between any observation and itself?
Sill \(s\): The value of variance that a variogram model attains at a given range (Figure 9.4).
\[ s=c_0+c_1 \]
Partial sill \(c_1\): The sill minus the nugget.
\[ c_1=s-c_0 \]
Partial sill to total sill ratio: The structural variance explained by the fitted variogram model (Rossi et al. 1992). This is the amount of variance that is spatially autocorrelated (Rossi et al. 1992).
\[ Ratio =\frac {s}{s+a}\]
Usually, we are interested in modeling the semivariance (semivariogram) or variance (variogram) of a process so that we can make predictions from a model rather than an incomplete set of sampled observations. In this way, we can fit a model to our paired observations and use a continuous variogram function to estimate the variance at any lag. These models are known as theoretical variograms and in the following sections we will compare several commonly-used models, summarized in Figure 9.5.
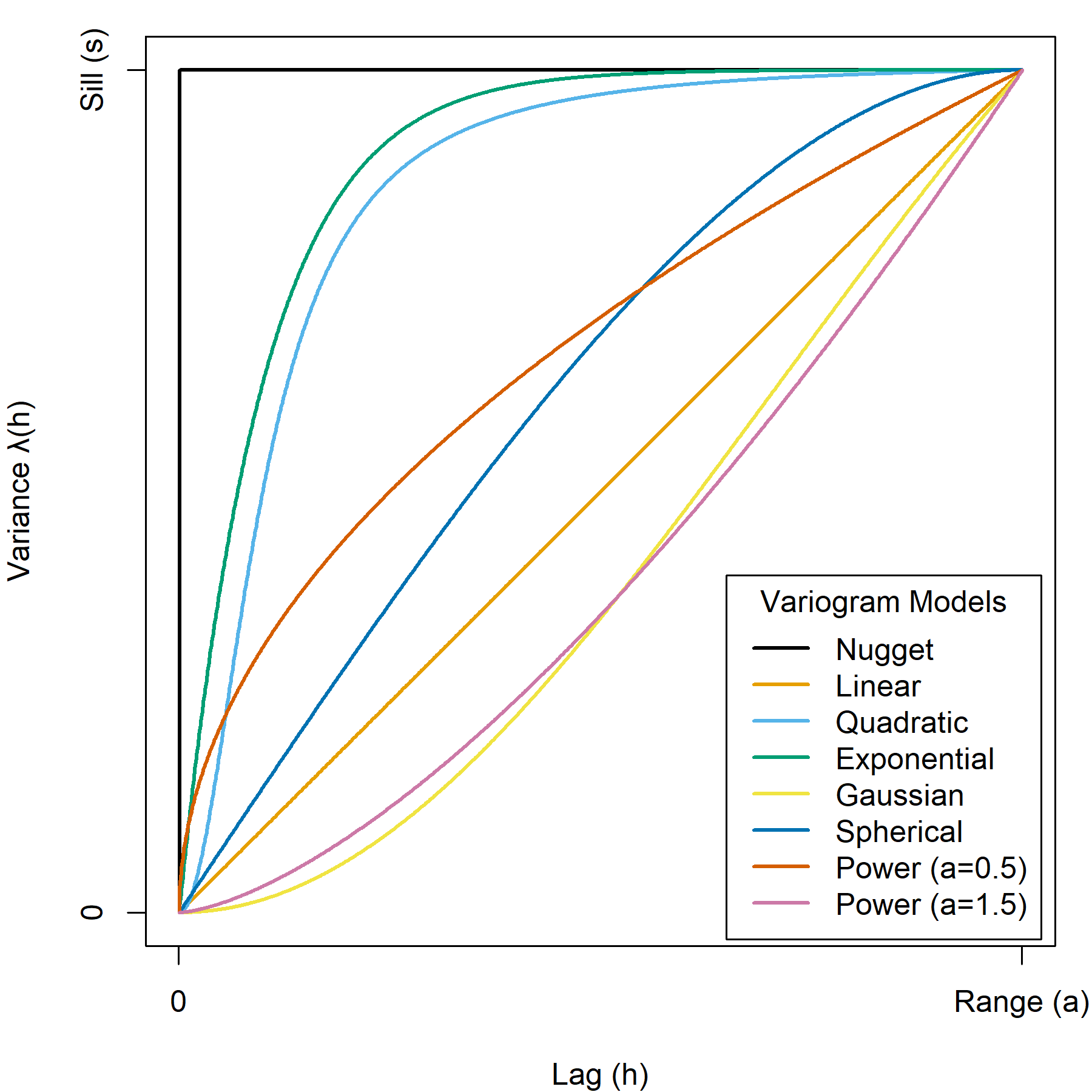
Figure 9.5: Comparison of theoretical variogram models. The upper bound of the variance in the plot is the sill for variogram models that have sills and the upper limit of the lag in the plot is the range for variogram models that have ranges. Note that the linear and power models are not bounded and extend infinitely beyond the plot space. The exponential, quadratic, and Gaussian models approach the sill asymptotically as lag distances approach infinity. Pickell, CC-BY-SA-4.0.
9.10.1 Nugget variogram model
A nugget model is the simplest model that assumes there is no relationship between variance and lag and consequently there is no range for this model. In other words, the variance is assumed to be constant. This is also the least likely case for most natural spatial phenomena. If you used this model to make spatial predictions, then your predictions would be made as if your data had no spatial component at all, so this is also the least useful model for spatial prediction. The nugget model is expressed as:
\[ \begin{array}{ccc} \gamma{(h)} = 0 & \text{for }h=0, \\ \gamma{(h)} = 1 & \text{ for }h>0 \end{array} \]
9.10.2 Linear variogram model
A linear model assumes a linear and constant slope \(b\) between variance and lag. Like the nugget model, the linear model has no defined range or sill. Unlike the linear model, variance is assumed to be infinite and therefore it is not possible to distinguish between correlated and uncorrelated lags. Note the special case of \(b=0\) is equivalent to the nugget model. A linear effect in spatial autocorrelation suggests a trend in your spatial data. The linear model is expressed as:
\[ \gamma{(h)} = bh+c_0 \] Where \(b\) is the slope to be estimated.
9.10.3 Quadratic variogram model
A quadratic or logistic model assumes variance increases near-exponentially, but the shape is more “S”-shaped or sigmoid. The quadratic model is expressed as:
\[ \gamma{(h)} = \frac{ah^2}{1+bh^2}+c_0 \] Where \(b\) is the slope to be estimated.
9.10.4 Exponential variogram model
An exponential model assumes spatial autocorrelation decreases exponentially with increasing lag distance. The variance in the exponential variogram approaches the sill asymptotically as \(h → ∞\), so unlike the spherical and nugget models, an exponential model never assumes a repeating variance beyond the range. Since the variance will continue to marginally increase with infinite lag distance, the range is defined as 95% of the sill \(a=0.95s\) or \(3a\) beyond which lag distances \(h\) are considered not spatially autocorrelated for the given variable. The exponential model is expressed as:
\[ \gamma{(h)} = c_1(1-e^{-\frac{h}{a}})+c_0 \]
9.10.5 Gaussian variogram model
A Gaussian model assumes that spatial autocorrelation is extremely high at short lag distances (i.e., variance is low) and then falls quickly towards zero (i.e., variance is high) at farther lag distances. This is a good model to use if you expect high local autocorrelation or phenomena that change rapidly with distance. The range is defined as 95% of the sill \(a=0.95s\) or \(\sqrt{3}a\) beyond which lag distances \(h\) are considered not spatially autocorrelated for the given variable. The Gaussian model is expressed as:
\[ \gamma{(h)} = c_1(1-e^{-(\frac{h}{a})^2})+c_0 \]
9.10.6 Spherical variogram model
A spherical model assumes a progressive, but not constant, decrease of spatial autocorrelation (i.e., increasing variance) until the range, beyond which autocorrelation is zero and variance remains the same. Note that the slope of a spherical model is generally higher (i.e., faster) than a linear model where \(h<a\). The spherical model is one of the most commonly used models because the assumption of zero autocorrelation beyond the range \(h>a\). The spherical model is expressed as:
\[ \begin{array}{ccc} \gamma{(h)} = c_1(\frac{3}{2}\frac{h}{a}-\frac{1}{2}(\frac{h}{a})^3)+c_0 & \text{for }0<h≤a, \\ \gamma{(h)} = s = c_0 + c_1 = 0 & \text{ for }h>a \end{array} \]
9.10.7 Power variogram model
A power model modulates the variance by raising the lag to a power \(λ\):
\[ \begin{array}{ccc} \gamma{(h)} = h^λ & \text{for }0<λ<2, \\ \gamma{(h)} = bh+c_0 & \text{for }λ=1 \end{array} \]
The power model has no range or sill that can be estimated and the variance process is considered to be infinite, the same as the exponential and Gaussian models. Notably, \(λ=1\) is equivalent to the linear model.
Case Study: Selecting the appropriate variogram model
We will look at some different models for estimating the semivariogram. We will use the ground plot data from the young stand monitoring (YSM) program data (Province of BC 2018) for Fort Saint Johns Timber Supply Area (TSA) in the province of British Columbia, Canada. Fort Saint Johns is divided into six blocks respectively Figure 9.6. There are a total of 108 YSM plots used in this study (Figure 9.6). The total basal area (\(m^2/ha\)) is the variable that we want to predict spatially. For each of the YSM plots, we will calculate the total basal area by adding the basal area for all trees within each plot. We will explore different variogram models with these data and check which model provides the best fit for the variogram.
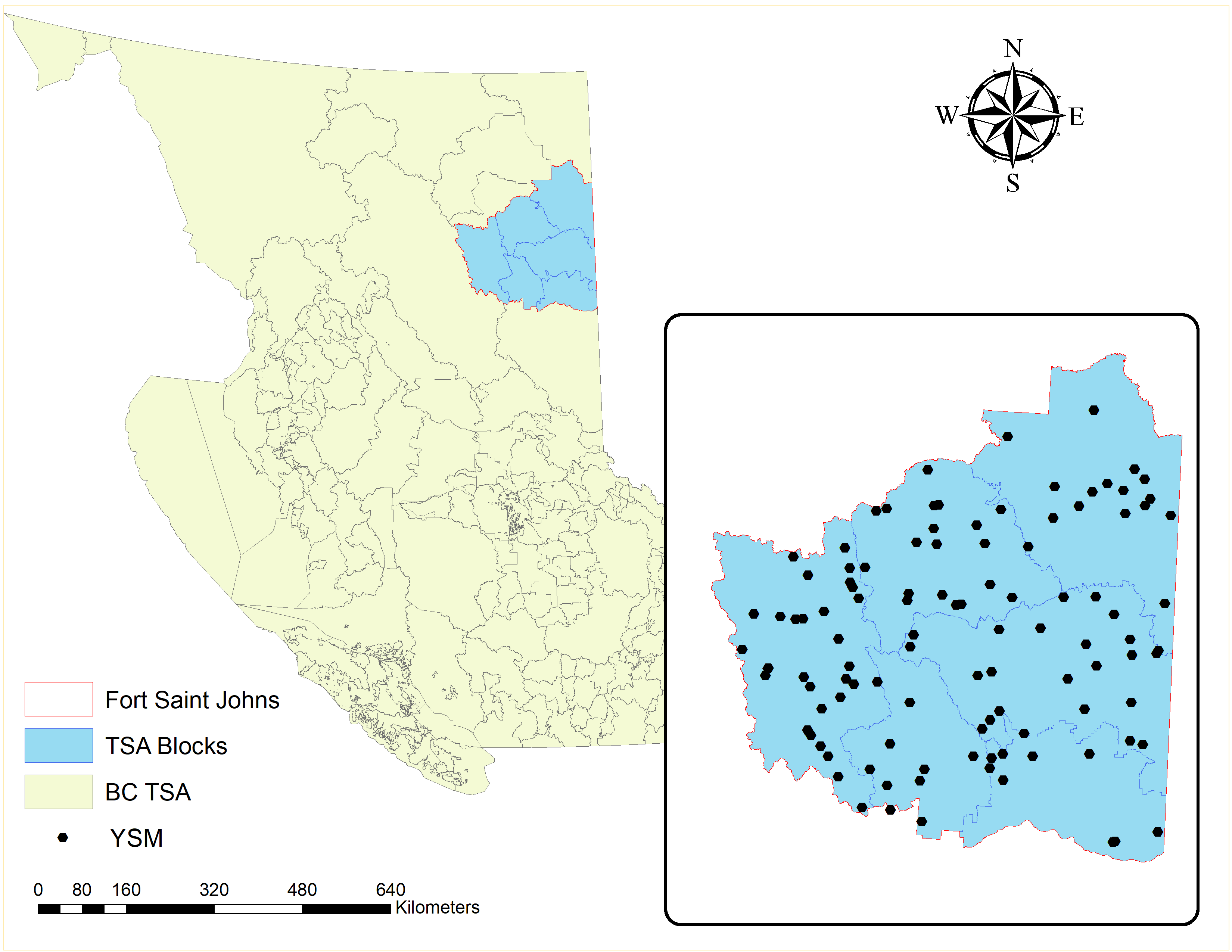
Figure 9.6: Location of Fort Saint Johns Timber Supply Area (TSA) in British Columbia (BC), Canada and the young stand monitoring (YSM) plots. Nepal, CC-BY-SA-4.0.

Figure 9.7: Semivariogram comparing different variogram models for basal area (m2/ha) of the young stand monitoring plots. Nepal, CC-BY-SA-4.0.
Just looking at the variograms, it appears that all of the four models fit our data well and indicates there is a strong correlation in basal area per hectare of live trees between the plots. However, we will use all the components of semivariogram models to pick our best fitting variogram.
| Model | Range | Nugget | Sill | Partial_sill | Sill_to_Sill |
|---|---|---|---|---|---|
| Circular | 7433.68 | 552.71 | 2338.27 | 1785.56 | 0.76 |
| Gaussian | 4446.62 | 0.00 | 2995.05 | 4446.62 | 1.00 |
| Spherical | 9729.37 | 14.13 | 2980.05 | 2966.37 | 0.99 |
| Exponential | 3871.76 | 0.00 | 2994.05 | 2994.05 | 1.00 |
We can see that the p-to-s ratio is highest for Gaussian and exponential semivariogram. This indicates the most total amount of semivariance that is spatially autocorrelated. Similarly, we can see that both models are indicating that we are observing a spatial autocorrelation in basal area at a very shorter range. Since with, exponential semivariogram autocorrelation only disappear at a infinite distance in reality, it is better to pick a Gaussian model in most of the similar cases like we have in this example.
9.11 Sampling
Sampling can be defined as the process of selecting some part of a population in order to make an inference, and estimate some parameters about the whole population(S. K. Thompson 2012). For example, to estimate the amount of biomass of trees in the University of British Columbia Malcolm Knapp Research Forest, scientists collect data on tree size and height from 100 randomly distributed small plots across the forest. Based on some allometric equations that relate these tree size measures to the mass of different species, biomass of the entire Malcolm Knapp Research Forest can be estimated. Similarly, to estimate the amount of recoverable oil in a region, a few (highly expensive) sample holes can be drilled (example adapted from S. K. Thompson (2012)). The situation is similar in a national opinion survey, in which only a sample of the people in the population are contacted, and the opinions in the sample are used to estimate the proportions with the various opinions in the whole population (example adapted from S. K. Thompson (2012)).
Sampling should not be confused with observational study. In an observational study, one has little-to-no control over the inclusion of units in the study whereas sampling usually consists of a well-defined protocol for defining both the population under study and the inclusion criteria of units to sample (S. K. Thompson 2012). Broadly, sampling can be categorized into two groups (Teddlie and Yu 2007):
- Probability sampling
- Non-probability sampling
Before getting into the details about different types of sampling. We will make ourself familiar with some sampling key terms and their definitions.
9.12 Population
Any large spatially defined entity of plots, people, trees, animals etc., from which samples are drawn and measurement of certain characteristics is conducted.
9.15 Probability Sampling
Probability sampling techniques are mostly used in studies that use extensive amount of quantitative analysis (Tashakkori and Teddlie 2010). It involves selecting a large number of units from a population where the probability of inclusion for every member of the population is determinable (Tashakkori and Teddlie 2010).
9.16 Simple Random Sampling
In simple random sampling, each sampling unit within a given population has equal probability of being selected in a sample (S. K. Thompson 2012).For example, suppose we would like to measure the tree heights of all the trees from a simple random sample of 60 plots with their spatial locations (given by plots center co-ordinates) from a forest divided into 625 spatially defined plots as given in Figure 9.8. Notice, there is no distinctive pattern on how plots are being selected for the measurement of tree heights, this justify the random part of the simple random sampling.
As an investigator, when we make a sequence of selections from a population, at each step, new and distinct set of sampling units are being selected in the sample, each having equal probability of being selected at each step.
For example, when we take another sample of 60 plots, we can see that different sampling units (plots) are being selected from what we obtained, this represent the equal probability of each sampling unit being selected.

Figure 9.8: Simple random sample of 60 units from a population of 625 units. Nepal, CC-BY-SA-4.0.
Simple random sample of 60 units from a population of 625 units.

Figure 9.9: Another simple random sample of 60 units. Nepal, CC-BY-SA-4.0.
9.17 Stratified Random Sampling
When a population under study is not homogeneous (similar in biological characteristics) across the entire study area and consists of some sort of gradient, stratified random sampling method is used (S. K. Thompson 2012). The principle of stratification is to partition the population in such a way that the units within a stratum are as similar as possible (Teddlie and Yu 2007). Random samples from each strata are drawn to ensure adequate sampling of all groups (Teddlie and Yu 2007). Even though one stratum may differ markedly from another, a stratified sample with the desired number of units from each stratum in the population will tend to be “representative” of the population as a whole (Howell et al. 2020).
For example, a forest under study is divided (stratified) into similar regions Figure 9.9 defined by elevation, soil moisture, and soil nutrient gradient and random samples are taken within each strata. The stratification of a study region despite of its size can help to spread the sample over the entire study area.

Figure 9.10: Stratified random sample within unequal strata within a study area. Nepal, CC-BY-SA-4.0.
Stratified random sample within unequal strata within a study area.

Figure 9.11: Stratified random sample from equal strata within a study area. Nepal, CC-BY-SA-4.0.
Stratified random sample from equal strata within a study area.
9.18 Systematic Sampling
A systematic sample uses a fixed grid or array to assign plots in a regular pattern Figure 9.11 (McRoberts, Tomppo, and Czaplewski 2014). The advantage of systematic sampling is that it maximizes the average distance between the plots and therefore minimizes spatial correlation among observations and increases statistical efficiency (McRoberts, Tomppo, and Czaplewski 2014). In addition, a systematic sample, which is clearly seen to be representative in some sense, can be very convincing to decision-makers who lack experience with sampling (McRoberts, Tomppo, and Czaplewski 2014). Raster grids such as digital elevation models (DEM) are some examples of systematic sample.

Figure 9.12: Sample every second observation in the row. Nepal, CC-BY-SA-4.0.
Sample every second observation in the row

Figure 9.13: Sample all the observation in every second column. Nepal, CC-BY-SA-4.0.
Sample all the observation in every second column
9.19 Cluster Sampling
In cluster sampling, rather than sampling individual units, which might be geographically spread over great distances, we can sample groups (clusters) of plots that occur naturally in the study area (Teddlie and Yu 2007). Cluster sampling is employed when we want to be more efficient in terms of the use of time and money to generate a more efficient probability sample (Teddlie and Yu 2007).

Figure 9.14: Cluster of plots selected from entire study area. Nepal, CC-BY-SA-4.0.
Cluster of plots selected from entire study area
9.20 Non-probability Sampling
Non-probability sampling is generally used in qualitative studies. They are also know as purposive or adaptive sampling, and defined as selecting units (e.g.,individuals, groups of individuals, institutions) based on specific purposes associated with answering some research questions. Purposive or Adaptive sampling can be classified into three broad categories (Teddlie and Yu 2007):
9.21 Representative Sampling
This type of sampling is used when we want to select samples that will represent broader groups as closely as possible (Teddlie and Yu 2007). One of the example of representative sampling is selecting 100 Douglas fir and 50 Spruce tree from study area within Malcom Knapp Forest, BC consisting of 500 Douglas fir and 300 Spruce trees for the measurement of tree height.
9.22 Unique Case Sampling
In this sampling, we want to focus on more specific case which is unique and rare in terms of one or more characteristics (Teddlie and Yu 2007). One of the example of unique case sampling could be understanding the genetic makeup of person who is not affected by Covid-19 virus.
9.23 Sequential Sampling
In this sampling method, we would pick up a single or group of cases in an interval of time, analyzes the results and then move on to the next group of cases and so on (Teddlie and Yu 2007). The goal of the research project is to generate some theory (or broadly defined themes) (Teddlie and Yu 2007).
9.24 Spatial Interpolation
Spatial interpolation can be defined as the process of predicting the given variable of interest at an unmeasured location given we have the sample in the proximity of the unknown location. Spatial interpolation methods can be categorized into two broad groups:
- Methods without using semivariogram
- Methods using semivariogram
We will discuss both method with a case study in detail.
9.25 Case Study 3
For this case study, we will use ground plot data from Young stand monitoring (YSM) program data (Province of BC 2018) for Fort Saint Johns timber supply area (TSA) in the province of British Columbia, Canada. Fort Saint Johns is divided into 6 blocks respectively Figure 9.6. There are a total of 108 YSM plots used in this study Figure 9.6. The total basal area (m2/ha) is our variable of interest in this study. For each of the YSM plot the total basal area was calculated by adding the basal area for all trees within the plot. We will use this dataset to explore different interpolation technique to find the variable of interest (basal area) within the unsampled locations.

Figure 9.15: Location of Fort Saint Johns TSA and the young stand change monitoring plots. Nepal, CC-BY-SA-4.0.
9.27 Nearest Neighbor
Nearest neighbor interpolation approach uses the value of variable of interest from the nearest sampled location and assign the value to the unsampled location of interest (Titus and Geroge 2013). It is very simple method and is most widely used for image processing in remote sensing research (Titus and Geroge 2013).
Step 1:Match the projection of plot data with the study area boundary
# spatstat Used for the dirichlet tessellation function
# maptools Used for conversion from SPDF to ppp
# raster Used to clip out thiessen polygons
spdf <- as_Spatial(filename)
dsp <- SpatialPoints(plot1[,14:15], proj4string=CRS("+proj=utm +zone=10 +ellps=GRS80 +datum=NAD83"))
dsp <- SpatialPointsDataFrame(dsp, plot1)
###############
TA <- CRS("+proj=utm +zone=10 +ellps=GRS80 +datum=NAD83")
library(rgdal)
dta <- spTransform(dsp, TA)
cata <- spTransform(spdf, TA)Step 2: Create polygons throughout the study area where interpolation is to be done and rasterize the polygons
#fig_cap <- paste0("An intermediate step in creating polygon and rasterizing it over the entire Fort Saint Johns TSA. Nepal, CC-BY-SA-4.0.")
#v <- voronoi(dta)
#plot(v)
### FUNCTION BELOW CRASHES R, COMMENTING OUT FOR NOW
#vca <- intersect(v, cata)
#spplot(vca, 'baha_L', col.regions=rev(get_col_regions()))
################### rasterize the data ####################
#r <- raster(cata, res=100)
#vr <- rasterize(vca, r, 'baha_L')An intermediate step in creating polygon and rasterizing it over the entire Fort Saint Johns TSA.
Step 3: Nearest neighbor with five unsampled points to be interpolated at a time and plot the results
### COMMENTING BELOW BECAUSE ABOVE CHUNK FAILS
#fig_cap <- paste0("Predicted basal area over the entire Fort Saint Johns TSA using five nearest neighbor. Nepal, CC-BY-SA-4.0.")
## gstat package to create semivariogram model, kriging an dinterpolation
#gs <- gstat(formula=baha_L~1, locations=dta, nmax=5, set=list(idp = 0))
#nn <- interpolate(r, gs)
#nnmsk <- mask(nn,vr)
#tm_shape(nnmsk) +
# tm_raster(n=8,palette = "RdBu", auto.palette.mapping = FALSE,title="Predicted basal area") +
# tm_legend(legend.outside=FALSE)Predicted basal area over the entire Fort Saint Johns TSA using five nearest neighbor.
Step 4: Leaflet map for some interactions
Predicted basal area over the entire Fort Saint Johns TSA using five nearest neighbor projected over the province of British Columbia.
9.28 Thiessian Polygon
In this method, the domain is determined into the area/polygons of regions containing one sampling point from the original data (Coulston and Reams, n.d.).The thiessen polygons are assigned with the same values of the variable of interest as the point sampled (Yamada 2016).
Step 1: Match the projection of the Shape file and the plot data
# project the plot data based to UTM zone 10 and NAD83
dsp <- SpatialPoints(plot1[,14:15], proj4string=CRS("+proj=utm +zone=10 +ellps=GRS80 +datum=NAD83"))
# convert the data into spatial object
dsp <- SpatialPointsDataFrame(dsp, plot1)
## change the projection of both shape file and plot data
TA <- CRS("+proj=utm +zone=10 +ellps=GRS80 +datum=NAD83")
library(rgdal)
dta <- spTransform(dsp, TA)
cata <- spTransform(spdf, TA)Step 2: Create the thiessian polygon around the sample points for entire TSA using
#"dirichlet" function from "spatstat" package
# Create a tessellated surface
th <- as(dirichlet(as.ppp(dta)), "SpatialPolygons")
# The dirichlet function does not carry over projection information
# requiring that this information be added manually to the thiessian polygons
proj4string(th) <- proj4string(dta)Step 3: The tessellated surface does not store attribute information from the point data layer. Hence, the information from the data layer should be carried over to tessellated surface
#We'll use the over() function from the "sp" package to join the point attributes to the thiessian polygon via spatial join
th.z <- over(th,dta, fn=mean)
th.spdf <- SpatialPolygonsDataFrame(th, th.z)
# Finally, we'll clip the tessellated surface to the Texas boundaries
th.clp <- raster::intersect(cata,th.spdf)Step 4: Visualize the results
fig_cap <- paste0("Predicted basal area over the entire Fort Saint Johns TSA using thiessan polygons. Nepal, CC-BY-SA-4.0.")
# Map the data
#using package "tmap"
tm_shape(th.clp) +
tm_polygons(col="baha_L", palette="RdBu", auto.palette.mapping = FALSE,title="Predicted basal area") +
tm_legend(legend.outside=FALSE)
Figure 9.16: Predicted basal area over the entire Fort Saint Johns TSA using thiessan polygons. Nepal, CC-BY-SA-4.0.
Inverse Distance Weighting
*Inverse distance weighting (IWD) (Shepard 1968) estimates the variable of interest by assigning more weight to closer points using the weighting function (w) based on the weighting exponent know as power (p) (Babak and Deutsch 2009). The influence of one data point on the other decreases as the distance increases. Hence, higher power of the exponent will result in point of interest having less effect on the points far from it (Babak and Deutsch 2009). It is a simple technique that does not require prior information to be applied to spatial prediction (Shepard 1968). Lower value of exponents mean more averaging, and the weights are more evenly distributed among the surrounding data points (Shepard 1968).
Step 1: Fix the projections between data points and shape file
# project the data based on the colorado plateau boundry projection
dsp <- SpatialPoints(plot1[,14:15], proj4string=CRS("+proj=utm +zone=10 +ellps=GRS80 +datum=NAD83"))
# convert the data into spatial object
dsp <- SpatialPointsDataFrame(dsp, plot1)
TA <- CRS("+proj=utm +zone=10 +ellps=GRS80 +datum=NAD83")
library(rgdal)
dta <- spTransform(dsp, TA)
cata <- spTransform(spdf, TA)Step 2: Create empty grid or over-lay empty grid over the study area
An empty grid over Fort Saint Johns TSA is created, where n is the total number of cells over which interpolation is to be done.
grd <- as.data.frame(spsample(dta, "regular", n=5000))
names(grd) <- c("X", "Y")
coordinates(grd) <- c("X", "Y")
gridded(grd) <- TRUE # Create SpatialPixel object
fullgrid(grd) <- TRUE # Create SpatialGrid object
# Add P's projection information to the empty grid
proj4string(dta) <- proj4string(dta) # Temp fix until new proj env is adopted
proj4string(grd) <- proj4string(dta)Step 3: Interpolate the grid cells using a power value of 2
Power values can be adjusted depending on characteristics of variable being interpolated
## [inverse distance weighted interpolation]Step 4: Plot the results from inverse distance weighting interpolation using
fig_cap <- paste0("Predicted basal area over the entire Fort Saint Johns TSA using inverse distance weighting. Nepal, CC-BY-SA-4.0.")
tm_shape(r.m) +
tm_raster(n=8,palette = "RdBu", auto.palette.mapping = FALSE,
title="Predicted basal area") +
tm_shape(dta) + tm_dots(size=0.2) +
tm_legend(legend.outside=FALSE)
Figure 9.17: Predicted basal area over the entire Fort Saint Johns TSA using inverse distance weighting. Nepal, CC-BY-SA-4.0.
Predicted basal area over the entire Fort Saint Johns TSA using inverse distance weighting.
Step 5: Leaflet map for some interaction
Figure 9.18: Predicted basal area over the entire Fort Saint Johns TSA using inverse distance weighting projected over the province of Brithish Columbia. Nepal, CC-BY-SA-4.0.
Figure 10.23. \(\ref{fig:figs}\) Predicted basal area over the entire Fort Saint Johns TSA using inverse distance weighting projected over the province of British Columbia.
9.30 Kriging
The spatial interpolation technique such as inverse distance weighting(IWD), nearest neighbor, and polygon approach are based on the surrounding neighborhood. There is another group of interpolation methods generally know as kriging (Krige 1951) which is based on both surrounding neighborhood and statistical models, especially spatial autocorrelation. Kriging uses the variogram modeling approach we studied in section 10.3 as a statistical model and incorporates the information about spatial autocorrelation while performing the interpolation. Since kriging uses the geostatistical model it has capacity of both prediction and provides some measure of the accuracy of prediction (Goovaerts 2008).The basic assumption of kriging is that the distance based samples reflect some degree of spatial correlation (Goovaerts 2008). We should note one thing that kriging works with raster surfaces where variable of interest are to be interpolated using the sampled locations. Kriging works with the following basic mathematical model: \[\hat{Z_{s_0}}=\sum_{i}^N \lambda_{i}Z_{s_i}\]
Where, \[\hat{Z_{s_o}}=\text{variable of interest to predicted at unsampled loaction}\ s_0 \] \[ \lambda=\text{an unknown value of weight at the measured } s_{i} location\] \[Z_{s_i}=\text{measured value at the sampled location} \ s_i\]
The goal of kriging is to determine the weights \[\lambda_i\] that will minimize the variance estimator of the predicted value and actual value at the unsampled location:
\[ Var|\hat{Z}_{s_o}-Z_{s_o}|\]
The \[\hat{Z}_{s_0}\] is decomposed into a trend component \[\mu_{s_o}\], which is the mean function as seen in the following equation:
\[\hat{Z}_{s_o} = \mu_{s_o} + \epsilon_{s_o}\]
Where,
\[\epsilon_{s_o} = \text{spatially autocorrelated erros}\]
9.30.1 Linear Kriging
Linear kriging are distribution free linear interpolation techniques that are in alignment with linear regression methods (Asa et al. 2012). There are three principle linear kriging techniques as discussed below:
9.31 Case Study 4
For this case study, we will use ground plot data from Young stand monitoring (YSM) program data (Province of BC 2018) for Fort Saint Johns timber supply area (TSA) in the province of British Columbia, Canada. Fort Saint Johns is divided into 6 blocks respectively Figure 9.6. There are a total of 108 YSM plots used in this study Figure 9.6. The total basal area (m2/ha) is our variable of interest in this study. For each of the YSM plot the total basal area was calculated by adding the basal area for all trees within the plot. We will use this dataset to explore different interpolation technique to find the variable of interest (basal area) in the unsampled locations.
9.32 Simple Kriging
Simple kriging works with the assumption that the mean is known and constant over entire domain and calculated as the average of the data (Wackernagel 2002). The number of sampled points used to make the prediction of the variable of interest in unmeasured location depends upon the range of semivariogram model used (Burrough and McDonnell 1998).
\[\hat{Z}_{s_o}=\mu_{s_o} + \epsilon_{s_o}\] Where, \[\hat{Z}_{s_o}=\text{variable of interest predicted at a given saptial location}\ s_{o}\] \[\mu_{s_o}=\text{an known constant mean} \]
Step 1: Make sure the projection of the point data and shape file is same
# summarize the data to individual plots
data<-plot1 %>%
group_by(utm_eastin,utm_northi) %>%
summarise(total= sum(baha_L))
# convert the data to saptial point data frame and change the projection to NAD83
dsp <- SpatialPoints(data[,1:2], proj4string=CRS("+proj=utm +zone=10 +ellps=GRS80 +datum=NAD83"))
dsp <- SpatialPointsDataFrame(dsp, data)
# Make the projection similar for plot and polygon data
TA <- CRS("+proj=utm +zone=10 +ellps=GRS80 +datum=NAD83")
dta <- spTransform(dsp, TA)
cata <- spTransform(spdf, TA)Step 2: Create an empty grid
A grid with the total number of n cells where basal area is to be predicted is overlaid over Forty Saint Johns. The grid will be a raster
grd <- as.data.frame(spsample(dta, "regular", n=10000))
names(grd) <- c("X", "Y")
coordinates(grd) <- c("X", "Y")
gridded(grd) <- TRUE # Create SpatialPixel object
fullgrid(grd) <- TRUE # Create SpatialGrid object
# Add projection information to the empty grid relative to the plot and polygon projection
proj4string(dta) <- proj4string(dta) # Temp fix until new proj env is adopted
proj4string(grd) <- proj4string(dta)Step 3: Calculate the overall mean of the variable to be interpolated
Simple kriging, which assumes that mean is a known constant over entire domain need a mean value of variable of interest (basal area)
## [1] 95.72555Step 4: Semivariogram modeling
Start by fitting the semivariogram model for the variable of interest (basal area) and see which model best fit the data
fig_cap <- paste0("Variogram models fitted for basal area using the YSM plot data. Nepal, CC-BY-SA-4.0.")
#### Exponential variogram ###############
TheVariogram=variogram(total~1, data=dta)
TheVariogramModel <- vgm(psill=2500, model="Exp", nugget=1500, range=20000)
FittedModel <- fit.variogram(TheVariogram, model=TheVariogramModel)
preds = variogramLine(FittedModel, maxdist = max(TheVariogram$dist))
g<-ggplot(TheVariogram,aes(x=dist,y=gamma))+geom_point()+
geom_line(data = preds)+ theme_classic()+
labs(x = "lag distance (h)", y = "Semi-variance")+
theme_bw()+ ggtitle("Exponential")+
theme(text = element_text(size =14))+
theme(axis.title.x=element_blank(),
axis.title.y=element_text(size=14),
axis.text.x =element_blank(),
axis.text.y =element_text(size=14))
#### Spherical Variogram
TheVariogramModel1 <- vgm(psill=2500, model="Sph", nugget=1500, range=20000)
FittedModel1 <- fit.variogram(TheVariogram, model=TheVariogramModel1)
preds1 = variogramLine(FittedModel1, maxdist = max(TheVariogram$dist))
h<-ggplot(TheVariogram,aes(x=dist,y=gamma))+geom_point()+
geom_line(data = preds1)+ theme_classic()+
labs(x = "lag distance (h)", y = "Semi-variance")+
theme_bw()+ggtitle("Spherical")+
theme(text = element_text(size =14))+
theme(axis.title.x=element_blank(),
axis.title.y=element_blank(),
axis.text.x =element_blank(),
axis.text.y =element_blank(),
axis.ticks.y= element_blank())
## Gaussian Variogram
TheVariogramModel2 <- vgm(psill=2500, model="Gau", nugget=1500, range=20000)
FittedModel2 <- fit.variogram(TheVariogram, model=TheVariogramModel2)
preds2 = variogramLine(FittedModel2, maxdist = max(TheVariogram$dist))
i<-ggplot(TheVariogram,aes(x=dist,y=gamma))+geom_point()+
geom_line(data = preds2)+ theme_classic()+
labs(x = "lag distance (h)", y = "Semi-variance")+
theme_bw()+ggtitle("Gaussian")+
theme(text = element_text(size =14))+
theme(axis.title.x=element_text(size=14),
axis.title.y=element_text(size=14),
axis.text.x =element_text(size =14),
axis.text.y =element_text(size=14))
## circular Variogram
TheVariogramModel3 <- vgm(psill=2500, model="Cir", nugget=1500, range=20000)
FittedModel3 <- fit.variogram(TheVariogram, model=TheVariogramModel3)
preds3 = variogramLine(FittedModel3, maxdist = max(TheVariogram$dist))
j<-ggplot(TheVariogram,aes(x=dist,y=gamma))+geom_point()+
geom_line(data = preds3)+ theme_classic()+
labs(x = "lag distance (h)", y = "Semi-variance")+
theme_bw()+ggtitle("Circular")+
theme(text = element_text(size =14))+
theme(axis.title.x=element_text(size=14),
axis.title.y=element_blank(),
axis.ticks = element_blank(),
axis.text.x =element_text(size =14),
axis.text.y =element_blank())
########### combine all the plots together #########
####################################################################################
grids_bs <- plot_grid(g,h,i,j,ncol=2,align = "h")
grids_bs
Figure 9.19: Variogram models fitted for basal area using the YSM plot data. Nepal, CC-BY-SA-4.0.
Variogram models fitted for basal area using the YSM plot data.
Step 5: Put all the variogram parameters in a table and see which fits best
Table 10.2 Summary of various components of variogram from four variogram models.
| Model | Range | Nugget | Sill | Partial_sill | Partial_sill_to_Sill |
|---|---|---|---|---|---|
| Exponential | 3872.10 | 0.00 | 2994.10 | 2994.10 | 1 |
| Spherical | 9704.83 | 0.00 | 2994.64 | 2994.64 | 1 |
| Gaussian | 4552.88 | 0.00 | 2995.03 | 2995.03 | 1 |
| Circular | 7801.57 | 729.25 | 2161.74 | 1432.49 | 0.66 |
Looking into the variogram and the parameter (Figure 9.19, Table: (#tab:10-simple-kriging-5), we can see that exponential variogram fits the data quite well compared to spherical and Gaussian as it has short range, low nugget and high partial sill to total sill ratio. As pointed earlier, the spatial autocorrelation only disappear at infinite lag using exponential model and best way to go is Gaussian for our data.
Step 6: We will use beta = 95.73, as we know the assume mean is a known constant over the domain for simple kriging
## [using simple kriging]Step 6: Visualize the predicted surface
fig_cap <- paste0("Predicted basal area using simple kriging. Nepal, CC-BY-SA-4.0.")
#convert the kriging results to raster
raster_krig <- raster(simple)
raster_clip <- mask(raster_krig, cata)
# Plot the kriging result using the library tmap
tm_shape(raster_clip) +
tm_raster(n=8,palette = "RdBu", auto.palette.mapping = FALSE,
title="Predicted basal area using simple kriging") +
tm_shape(dta) + tm_dots(size=0.2) +
tm_legend(legend.outside=FALSE)
Figure 9.20: Predicted basal area using simple kriging. Nepal, CC-BY-SA-4.0.
Predicted basal area using simple kriging.
Step 7: Cross validation
cv_sK <- krige.cv(total~ 1, dta, model=FittedModel2, nfold=nrow(dta),
verbose=FALSE)
### calculate RMSE
res <- as.data.frame(cv_sK)$residual
sqrt(mean(res^2))## [1] 60.42301## [1] -0.6926475#### Mean squared deviation of the prediction VS the sample
mean(res^2/as.data.frame(cv_sK)$var1.var)## [1] 5.2580139.33 Ordinary Kriging
Ordinary krigning (G. Matheron 1973) is one of the most widely used method of kriging which assume that the mean for variable of interest is an unknown constant within the domain. The mean is calculated based on the sample that is within the search window, i.e.,local mean instead of assumed constant mean over entire domain (I. Clark and Harper 2007) (Goovaerts 2008). It assumes the following model:
\[\hat{Z}_{s_o}=\mu_{s_o} + \epsilon_{s_o}\] Where, \[\hat{Z}_{s_o}=\text{variable of interest predicted at a given saptial location}\ s_{o}\]
\[\mu_{s_o}=\text{an unknown constant mean}\]
Steps 1, 3, 4, are similar to what we did for simple kriging and we don’t need step 2 as mean is assumed to be a unknown constant
Step 5: Using ordinary kriging
Krigging model is specified only using variable of interest.
## [using ordinary kriging]Step 6: Visualize the predicted surface
fig_cap <- paste0("Predicted basal area using ordinary kriging. Nepal, CC-BY-SA-4.0.")
#convert the kriging results to raster
raster_Ok <- raster(ordinary)
Ok_clip <- mask(raster_Ok, cata)
# Plot the kriging result using the library tmap
tm_shape(Ok_clip) +
tm_raster(n=8,palette = "RdBu", auto.palette.mapping = FALSE,
title="Predicted basal area using ordinary kriging") +
tm_shape(dta) + tm_dots(size=0.2) +
tm_legend(legend.outside=FALSE)
Figure 9.21: Predicted basal area using ordinary kriging. Nepal, CC-BY-SA-4.0.
Predicted basal area using ordinary kriging.
Step 7: Cross validation**
cv_oK <- krige.cv(total~ 1, dta, model=FittedModel2, nfold=nrow(dta),
verbose=FALSE)
### calculate RMSE
res <- as.data.frame(cv_oK)$residual
sqrt(mean(res^2))## [1] 60.42301## [1] -0.6926475#### Mean squared deviation of the prediction VS the sample
mean(res^2/as.data.frame(cv_oK)$var1.var)## [1] 5.2580139.34 Universal Kriging
The universal kriging is one of the variant of ordinary kriging. This method assume that the mean varies from location to location in a deterministic way (trend or drift) while the variance is constant throughout the domain (G. Matheron 1962). One example could be measurements of temperatures, which are commonly related to elevation (at a known rate of oC by m difference).
Step 1: This is similar to what we did for simple kriging and we will calculate the mean based on the drift or trend in data (more localized mean) using the spatial location of the plots
# Add X and Y to our original point dataframe, it is just how universal kriging formula takes the value of the co-ordinates.
dta$X <- coordinates(dta)[,1]
dta$Y<- coordinates(dta)[,2]
## We will model the trend or drift using the location as X and y
TheVariogram4=variogram(total~X+Y, data=dta)
TheVariogramModel4 <- vgm(psill=2500, model="Gau", nugget=1500, range=20000)
FittedModel4 <- fit.variogram(TheVariogram4, model=TheVariogramModel4)
preds4 = variogramLine(FittedModel4, maxdist = max(TheVariogram4$dist))
g<-ggplot(TheVariogram4,aes(x=dist,y=gamma))+geom_point()+
geom_line(data = preds4)+ theme_classic()+
labs(x = "lag distance (h)", y = "Semi-variance")+
theme_bw()+ ggtitle("Gaussian")+
theme(text = element_text(size =14))+
theme(axis.title.x=element_blank(),
axis.title.y=element_text(size=14),
axis.text.x =element_blank(),
axis.text.y =element_text(size=14))
## using locations of the sample to calculate the localized mean
universal<- krige(total~X+Y,dta, grd, model=FittedModel4)## [using universal kriging]Step 2: Visualize the results as we usually
fig_cap <- paste0("Predicted basal area using universal kriging. Nepal, CC-BY-SA-4.0.")
#convert the kriging results to raster
raster_uk <- raster(universal)
uk_clip <- mask(raster_uk, cata)
# Plot the kriging result using the library tmap
tm_shape(uk_clip) +
tm_raster(n=8,palette = "RdBu", auto.palette.mapping = FALSE,
title="Predicted basal area using universal kriging") +
tm_shape(dta) + tm_dots(size=0.2) +
tm_legend(legend.outside=FALSE)
Figure 9.22: Predicted basal area using universal kriging. Nepal, CC-BY-SA-4.0.
Predicted basal area using universal kriging.
Step 3: Cross validation
cv_uK <- krige.cv(total~ X+Y, dta, model=FittedModel4, nfold=nrow(dta),
verbose=FALSE)
### calculate RMSE
res <- as.data.frame(cv_uK)$residual
sqrt(mean(res^2))## [1] 68.48368## [1] -1.374287#### Mean squared deviation of the prediction VS the sample
mean(res^2/as.data.frame(cv_uK)$var1.var)## [1] 10.97501Which method is the best given our data?
We will put the cross validation results together and cross compare across three methods.
Table 10.3 Cross-validation results for different type of kriging.
| Method | RMSE | ME | MSDR |
|---|---|---|---|
| Simple | 60.42 | -0.69 | 5.25 |
| Ordinary | 60.42 | -0.69 | 5.25 |
| Universal | 68.48 | -1.37 | 10.97 |
From the cross validation results (Table 10.3), we want root mean squared error (RMSE) to be low for greater predictive accuracy (Tziachris et al. 2017). We also want mean error (ME) to be as close to 0 as possible (Tziachris et al. 2017). And we want mean squared deviation Ratio (MSDR) to be closer to 1 for the good kriging model (Tziachris et al. 2017). The RMSE is lowest for simple and ordinary kriging, the ME is negative and below zero, and the MSDR of the predictions vs. the sample is low and closer to 1 for both simple and ordinary kriging (Table 10.3). This means the variability in predictions from both kriging are somewhat closer to real values than universal kriging (Tziachris et al. 2017). Looks like universal kriging is predicting more negative basal area for some of the location within TSA. The choice between simple and ordinary kriging may vary with researchers discretion here.
9.35 Co-Kriging
Co-kriging uses the information about different covariates to predict the value of the variable of interest at an unsampled location (Cressie 1994). It utilizes the autocorrelation of the variable of interest and the cross correlations between the variable of interest with all the covariates to make the prediction (Cressie 1994). In order to implement co-kriging we need to have a strong correlation between the covariates (Tziachris et al. 2017). The spatial variability of one variable should be correlated with the spatial variability of the other covariates (Tziachris et al. 2017).
9.36 Non-Linear Kriging
In principle, nonlinear kriging algorithms are linear kriging algorithms applied to nonlinear transformations of the data points into a continuous variable (Deutsch and Journel 1993). We will briefly talk about four principal non-linear kriging techniques in this chapter while our focus mostly is on linear kriging methods.
9.37 Indicator Kriging
Indicator kriging is a non-parametric approach of estimating a binary variable (presence absence or variables that takes 0 or 1 value) of interest at an unsampled or unmeasured location (Journel 1983). For example, we might have a sample that consists of information on presence or absence of Douglas-fir tree species within Williams lake timber supply area, where 0 indicates absence and 1 indicates the presence of species. Indicator kriging assumes that mean is a unknown constant over the domain. The only difference between the indicator kriging and ordinary krigging is in the use of binary variable. The basic mathematical formulation of indicator kriging is given below:
\[I_{s}= \mu + \epsilon_{s}\] where, \[I= \text{binary variable preicated at the location s}\] \[\mu= \text{unknown mean}\] \[\epsilon_{s}=\text{spatially autocorrelated error}\]
9.38 Probability Kriging
Probability kriging is useful when the variable of interest is binary as in case of indicator kriging. It is a special form of co-kriging which estimate the conditional probability that the unknown value of a variable at an unsampled location is above a specified cutoff level (Carr and Mao 1993). As in co-kriging, this method utilizes the autocorrelation of variable of interest and the cross correlations between the variable of interest with all the covariates to make the prediction (Carr and Mao 1993)
9.39 Disjunctive Kriging
Disjunctive kriging allows to estimate the value of a variable of interest at an unsampled location and estimating the conditional probability that the unknown value of a variable at an unsampled location is above a specified cutoff level (Yates, Warrick, and Myers 1986). Disjunctive kriging transforms the data into a normal distribution and then determine the probability that true value of variable at each location of interest exceeds the predefined threshold or cut-off probability (Daya and Bejari 2015).
9.40 Spatial Regression Models
For classical statistics tests, spatial autocorrelation is problematic as ordinary least square (regression) or analysis of variance (ANOVA) assumes that observations are independent in space and time (Meng et al. 2009). However, geostatistical data violates the assumptions of independence, and using regression and ANOVA might inflate the significance of t and F statistics, when, in fact, they may not be significant at all (Meng et al. 2009). In that case one should try to improve the regression model by adding important auxiliary (independent variables that are associated or important in predicting the variables of interest) and incorporating the spatial autocorrelation structure (Meng et al. 2009) using spatial regression models (Anselin and Bera 1998). The whole objective of spatial regression is to understand the association of the variable of interest with the independent variables while accounting for the spatial structure present in the data. We will show two examples of spatial regression model in this section using our familiar YSM data for Fort Saint Johns TSA.
9.41 Case Study 5
For this case study, we will use ground plot data from Young stand monitoring (YSM) program data (Province of BC 2018) for Fort Saint Johns timber supply area (TSA) in the province of British Columbia, Canada. Fort Saint Johns is divided into 6 blocks respectively Figure 9.6. There are a total of 108 YSM plots used in this study Figure 9.6. The total basal area (\(m^2/ha\)) is our variable of interest or response variable in this study. For each of the YSM plot the total basal area was calculated by adding the basal area for all trees within the plot. We will use the auxiliary variables such as trees per hectare (TPH), elevation (m), site index, top height (m), and tree volume (\(m^3/ha\))
9.42 Spatial Lag Model
Spatial lag models assume that the spatial autocorrelation only exist in the response variable or the variable of interest (Anselin and Bera 1998). Spatial lag model has the following general mathematical formulations:
\[ y= (\rho)WY + \beta X + \epsilon\] where, \[y= \text{response variable or variable of interest}\] \[\rho= \text{coefficients for the spatial weight matrix W}\] \[\beta= \text{coefficients for the predictor variables}\] \[WY=\text{spatially lagged response variable for the weight matrix W}\] \[X=\text{matrix of observations for the predictor variables}\] \[\epsilon=\text{vector of error terms}\]
9.43 Steps in Fitting Spatial Lag Model:
Step 1: Build spatial weight matrix
We will create a spatial weight matrix using the distance-based approach using the nearest-neighbor approach. This spatial weight matrix will be used in our spatial lag model to account for the spatial autocorrelation.
Step 2: Check for the spatial autocorrelation
We will check the spatial autocorrelation using the Moran’s I and Moran’s test using our distance based spatial weight matrix (Getis and Aldstadt 2010) we have just calculated.
##
## Moran I test under randomisation
##
## data: YSM_pots$Basal
## weights: YSM.W
##
## Moran I statistic standard deviate = 2.6524, p-value = 0.003996
## alternative hypothesis: greater
## sample estimates:
## Moran I statistic Expectation Variance
## 0.0656590005 -0.0093457944 0.0007996302Moran’s I value was (I=0.065), which was significant (p-value < 0.05) indicting a positive spatial autocorrelation. It seems like a weak autocorrelation but for the purpose of demonstration in this section we will proceed further assuming there is a spatial autocorrelation.
Step 3: Fit a spatial lag model
In this step, we will fit a spatial lag model using basal area (m2/ha) as a response variable. While, trees per hectare (TPH), elevation (m), site index, top height(m), and tree volume (m3/ha) will be used as predictor variable.
##
## Call:lagsarlm(formula = f1, data = YSM_pots, listw = YSM.W, zero.policy = T)
##
## Residuals:
## Min 1Q Median 3Q Max
## -28.4814 -8.7285 -1.5817 8.6639 34.8502
##
## Type: lag
## Coefficients: (asymptotic standard errors)
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -2.0836e+01 1.2070e+01 -1.7262 0.0843163
## TPH 3.7555e-03 4.6693e-04 8.0430 8.882e-16
## Elevation 2.0548e-02 5.3747e-03 3.8230 0.0001318
## Volume 9.6251e-02 4.8825e-03 19.7135 < 2.2e-16
## Height -8.6348e-02 4.7058e-01 -0.1835 0.8544098
## Site_index -4.3118e-01 3.3104e-01 -1.3025 0.1927520
##
## Rho: 0.18185, LR test value: 5.015, p-value: 0.025129
## Asymptotic standard error: 0.079411
## z-value: 2.29, p-value: 0.022023
## Wald statistic: 5.244, p-value: 0.022023
##
## Log likelihood: -425.7183 for lag model
## ML residual variance (sigma squared): 155.08, (sigma: 12.453)
## Number of observations: 108
## Number of parameters estimated: 8
## AIC: 867.44, (AIC for lm: 870.45)
## LM test for residual autocorrelation
## test value: 0.82749, p-value: 0.363Step 4: Select auxiliary variables and refit the model
Varieties of ways has been proposed to select the auxiliary variables to get the best spatial model. We will go through them briefly. First, we will Use the alpha=0.005 to check whether our auxiliary variables are significantly associated with the basal area. From the summary, we can see that TPH, Elevation and Volume has p-value < 0.05 indicating that they are significantly associated with basal area.
##
## Call:lagsarlm(formula = f2, data = YSM_pots, listw = YSM.W, zero.policy = T)
##
## Residuals:
## Min 1Q Median 3Q Max
## -29.94846 -7.71629 -0.30949 8.76752 35.90219
##
## Type: lag
## Coefficients: (asymptotic standard errors)
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -2.8815e+01 9.0662e+00 -3.1783 0.001481
## TPH 3.8730e-03 4.1327e-04 9.3716 < 2.2e-16
## Elevation 2.4226e-02 4.5404e-03 5.3356 9.52e-08
## Volume 9.4008e-02 2.3532e-03 39.9498 < 2.2e-16
##
## Rho: 0.16445, LR test value: 4.1412, p-value: 0.041852
## Asymptotic standard error: 0.078353
## z-value: 2.0988, p-value: 0.035833
## Wald statistic: 4.405, p-value: 0.035833
##
## Log likelihood: -426.8261 for lag model
## ML residual variance (sigma squared): 158.35, (sigma: 12.584)
## Number of observations: 108
## Number of parameters estimated: 6
## AIC: 865.65, (AIC for lm: 867.79)
## LM test for residual autocorrelation
## test value: 1.3195, p-value: 0.25067Step 5: Assess both models using Akaike’s Information Criteria (AIC)
Sometime, only using the p-value to assess significant variables will not be useful while assessing which models best fits the data as multiple models can have potential to describe the association between response and predictor variables. In the context, when we have different competing models, we can use AIC (for details see, Akaike 1973) to compare the models. For example, suppose models we have fitted in step 3 and step 4 were competing and potential. We can select the best model between two with lowest AIC values. AIC value of model from step 3 is 867.44 while the AIC value of model from step 4 is 865.65, indicating later one is the best fit to our data.
Step 6: Interpreting rho coefficient for our selected model
Rho (0.16445), reflects the spatial dependence inherent in our sample data, measuring the average influence on observations by their neighboring observations. It has a positive effect and it is significant (p-value < 0.05). As a result, the general model fit improved over the linear model.
9.44 Spatial Error Model
Spatial error models assume that the spatial autocorrelation exists in the residuals or the error term of the regression equation (Anselin and Bera 1998). The general mathematical formula for spatial error model is given below:
\[ y=\beta X + \epsilon\] \[\epsilon= \lambda (W)\epsilon+ u\]
where, \[y= \text{response variable or variable of interest}\] \[\beta= \text{coefficients for the predictor variables}\] \[\lambda= \text{coefficients for the spatial weight matrix W for spatially autocorrelated erros}\]
\[(W)\epsilon=\text{spatial weight matrix W}\] \[X=\text{matrix of observations for the predictor variables}\] \[u=\text{indepndent errors}\]
9.45 Steps in Fitting Spatial Error Model:
Step 1 and Step 2 in fitting spatial error model is exactly similar to what we did for spatial lag model.
Step 3: Fit the spatial error model
We will use the function errorsarlm from the package spatialreg to fit the error model indicating that we are accounting the spatial autocorrelation that exists in the residuals or the error terms instead of the response variable (basal area).
##
## Call:errorsarlm(formula = f3, data = YSM_pots, listw = YSM.W, zero.policy = T)
##
## Residuals:
## Min 1Q Median 3Q Max
## -27.7331 -7.8589 -1.0210 7.8231 35.1950
##
## Type: error
## Coefficients: (asymptotic standard errors)
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -0.14171626 10.59980550 -0.0134 0.98933
## TPH 0.00378146 0.00048423 7.8092 5.773e-15
## Elevation 0.01635979 0.00645862 2.5330 0.01131
## Volume 0.09585359 0.00479585 19.9868 < 2.2e-16
## Height -0.11345603 0.45520593 -0.2492 0.80317
## Site_index -0.30051747 0.32907580 -0.9132 0.36113
##
## Lambda: 0.50793, LR test value: 3.6202, p-value: 0.057081
## Asymptotic standard error: 0.1857
## z-value: 2.7352, p-value: 0.0062338
## Wald statistic: 7.4815, p-value: 0.0062338
##
## Log likelihood: -426.4156 for error model
## ML residual variance (sigma squared): 154.69, (sigma: 12.438)
## Number of observations: 108
## Number of parameters estimated: 8
## AIC: NA (not available for weighted model), (AIC for lm: 870.45)Step 4: Select auxiliary variables and refit the model
We will use the alpha=0.05 to check whether our auxiliary variables are significantly associated with the basal area. From the summary, we can see that TPH, Elevation and Volume has p-value < 0.05 indicating that they are significantly associated with basal area.
##
## Call:errorsarlm(formula = f4, data = YSM_pots, listw = YSM.W, zero.policy = T)
##
## Residuals:
## Min 1Q Median 3Q Max
## -27.84609 -7.91303 -0.65425 7.82677 36.06371
##
## Type: error
## Coefficients: (asymptotic standard errors)
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -7.12704410 6.71512779 -1.0613 0.288535
## TPH 0.00391705 0.00042822 9.1472 < 2.2e-16
## Elevation 0.01843299 0.00595547 3.0951 0.001967
## Volume 0.09366580 0.00235054 39.8487 < 2.2e-16
##
## Lambda: 0.52491, LR test value: 3.7554, p-value: 0.052636
## Asymptotic standard error: 0.18121
## z-value: 2.8967, p-value: 0.0037711
## Wald statistic: 8.3909, p-value: 0.0037711
##
## Log likelihood: -427.019 for error model
## ML residual variance (sigma squared): 156.21, (sigma: 12.498)
## Number of observations: 108
## Number of parameters estimated: 6
## AIC: NA (not available for weighted model), (AIC for lm: 867.79)Step 5: Assess both models using akaike’s information criteria (AIC)
AIC value of model from step 3 is 868.83 while the AIC value of model from step 4 is 866.04, indicating later one is the best fit to our data.
Step 6: Interpret lambda parameter from the summary from our selected model
The lag error parameter Lambda for the model in step 4 is positive and significant (p-value < 0.05), indicating the need to control for spatial autocorrelation in the error
9.46 Selection Between Lag and Error Model
When it is not so clear theoretically that either of the spatial model works for our data, we can compare the model performance parameters: the AIC and Log likelihood. In our case, the spatial error model has lowest AIC and highest negative Log likelihood values. Hence, spatial lag model best fits our data.
When the question is which of the two models is better? This is an open question. The general advice is first to look for a theoretical basis to inform your choice. If there are strong substantive grounds for one model instead of the other, you should adopt it.